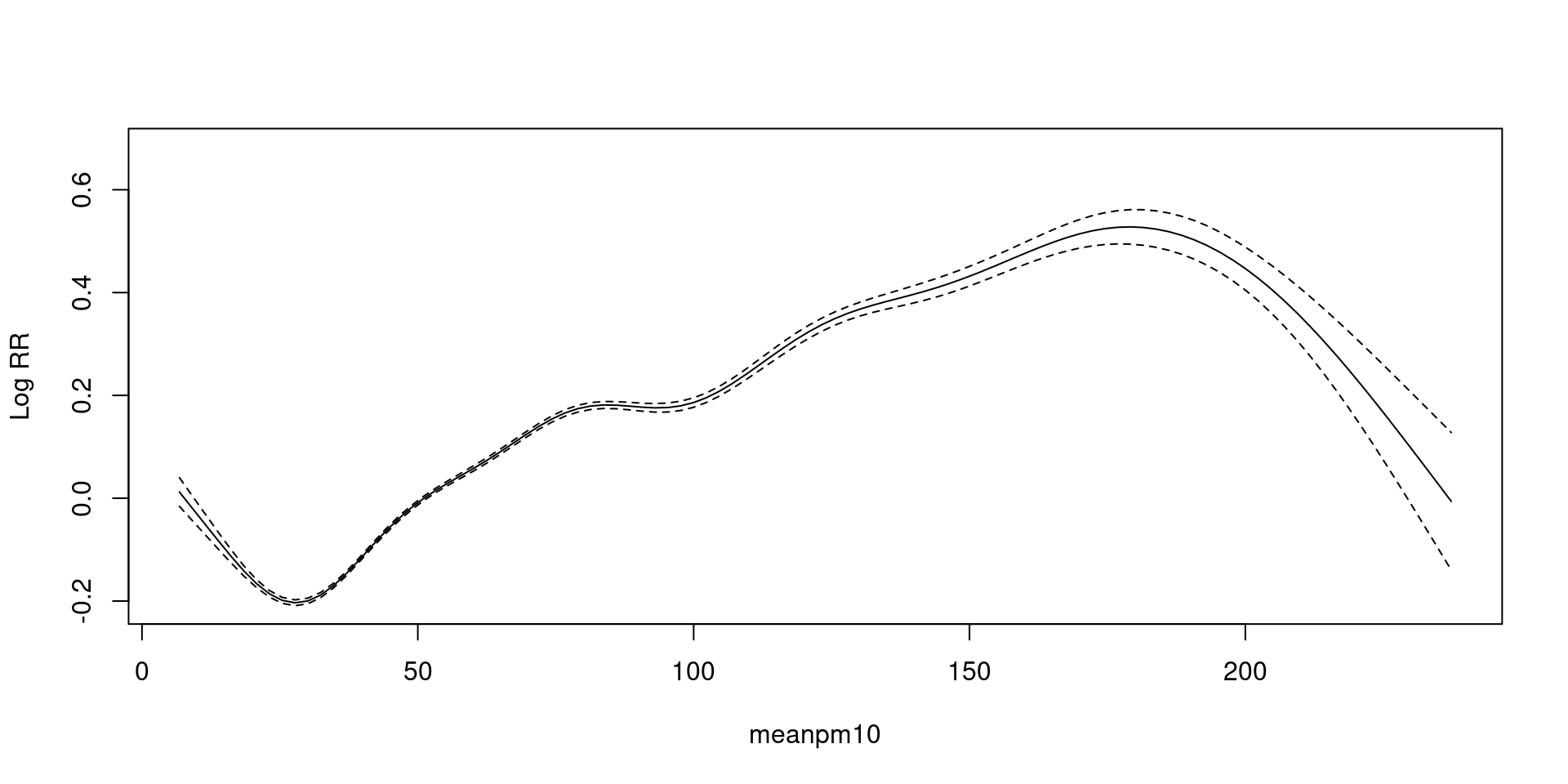
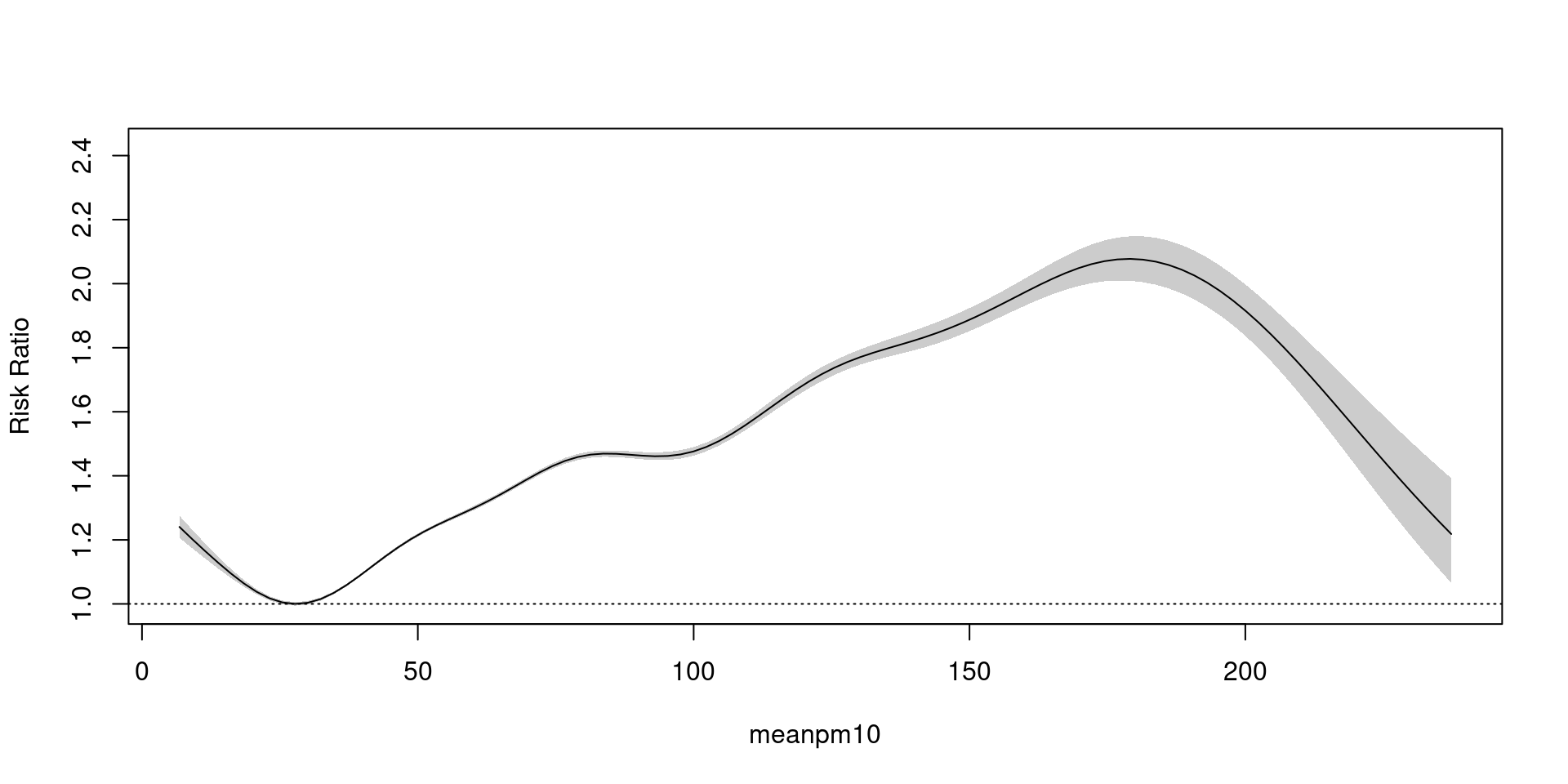
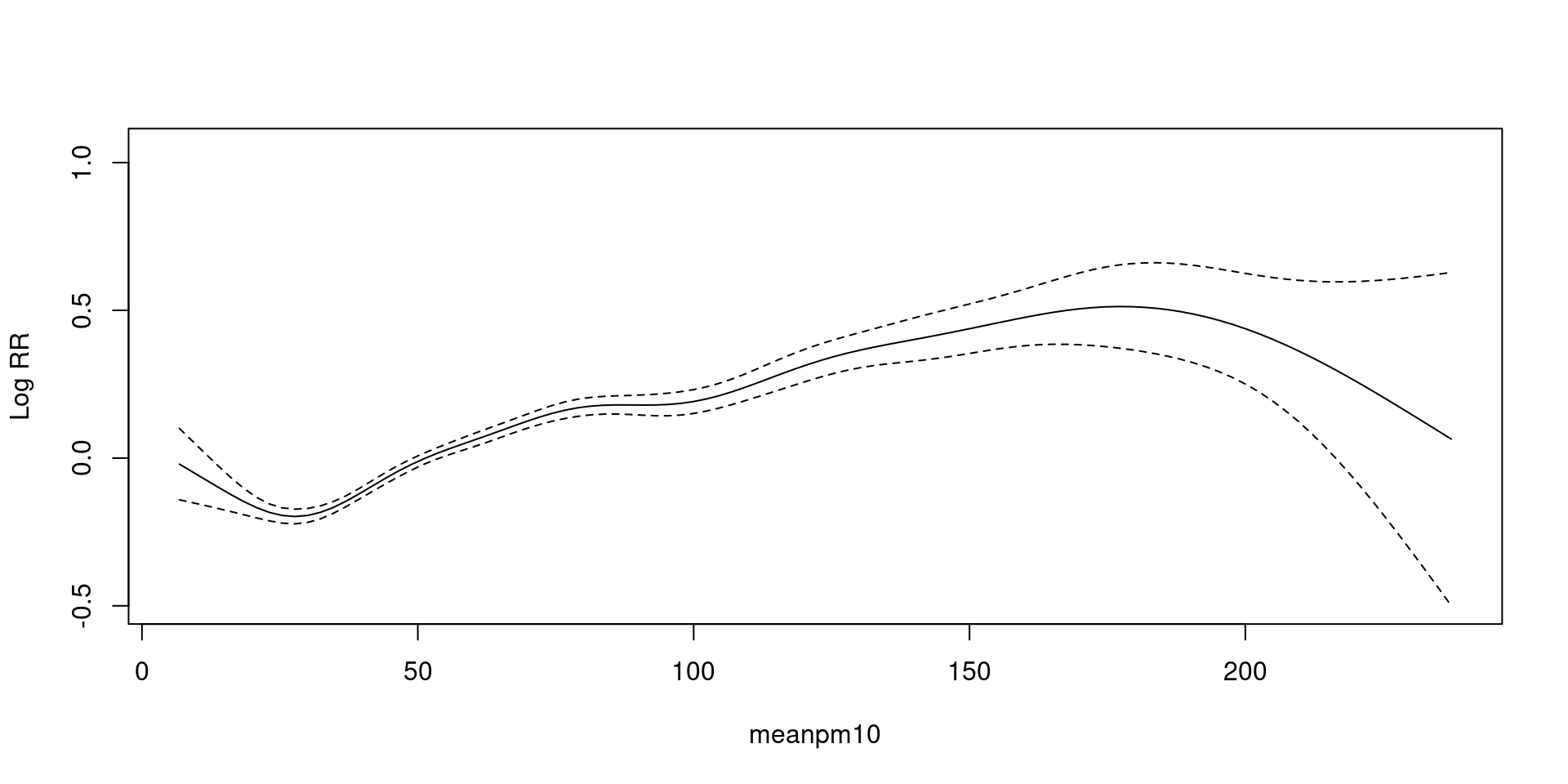
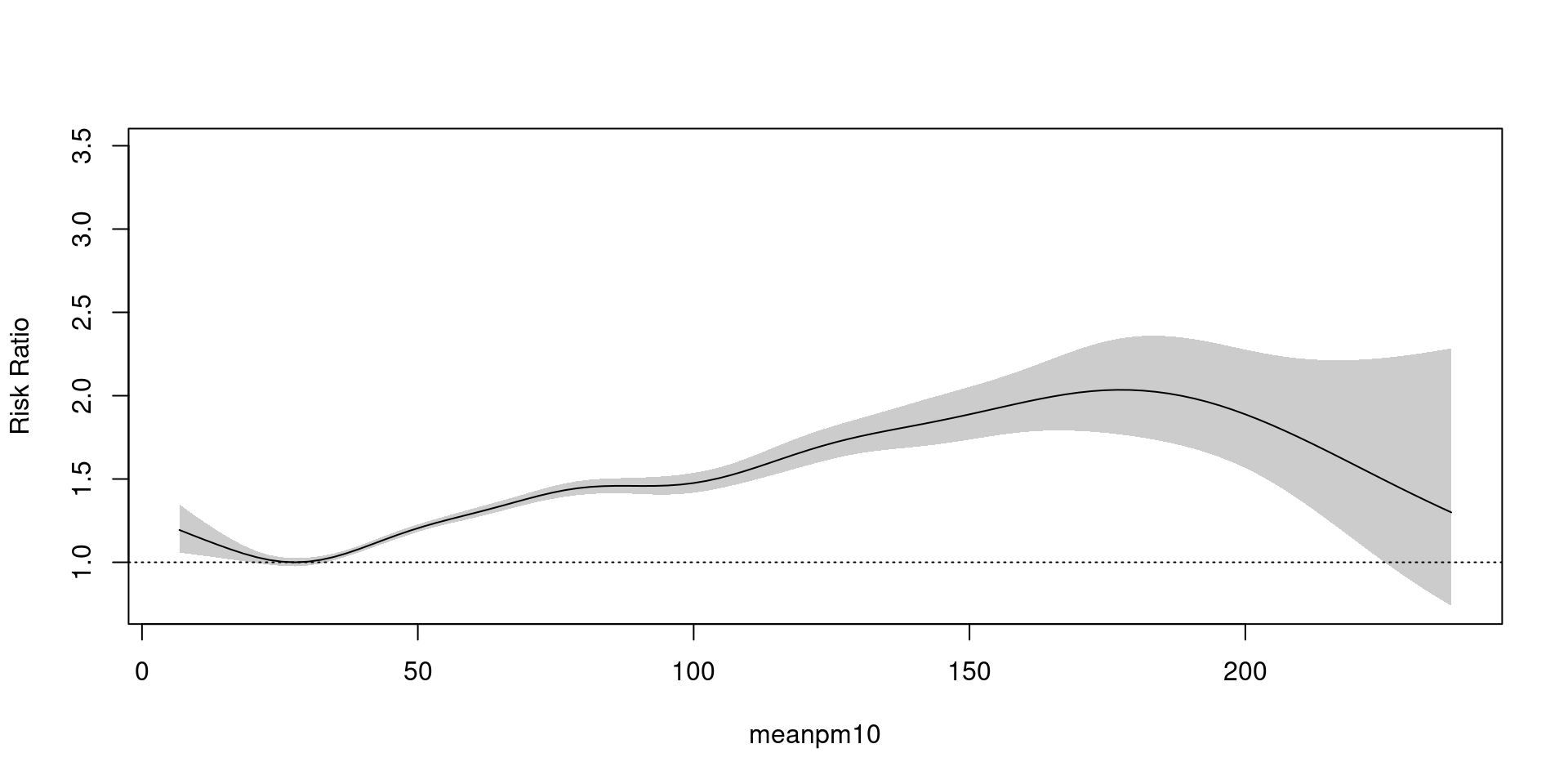
Generalized Additive Model(GAM)
Zarathu Co.,Ltd
Jinseob Kim
Executive summary
GAM 은 비선형관계를 다루는 통계방법이다
- LOWESS: 구간 촘촘하게 나눈 후 평균값
- Cubic spline(cs): 구간 몇개로 나눈 후 각각 3차함수 fitting
- Natural cubic spline(ns): cs 맨 처음과 끝구간만 선형 fitting
- Smoothing spline(GAM default): 최적화때 smoothing penalty(\(\lambda\)) 부여
종속변수 형태따라 여러종류
- Continuous: normal
- Binary: logistic
- Count: poisson, quasipoisson(평균 \(\neq\) 분산 일 때)
- Survival: coxph
GAM theory
Non linear model
\[\begin{align} Y=\beta_0+\beta_1 x_{1}+\beta_2 x_2+\cdots+\epsilon \end{align}\] \[\begin{align} Y=\beta_0+ f(x_1)+\beta_2 x_2 \cdots+\epsilon \end{align}\] \(f(x_1,x_2)\)꼴의 형태도 가능
LOWESS
Locally weighted scatterplot smoothing
- 구간 나눠 regression
- 각 점마다 그 점을 포함하는 구간 설정
- 가까운 점에 weight

LOWESS in ggplot

Cubic spline
Cubic = 3차방정식
- 구간을 몇 개로 나누고: knot
- 각각을 3차함수로 fitting
- 구간 사이 부드럽게 연결되도록 제한조건
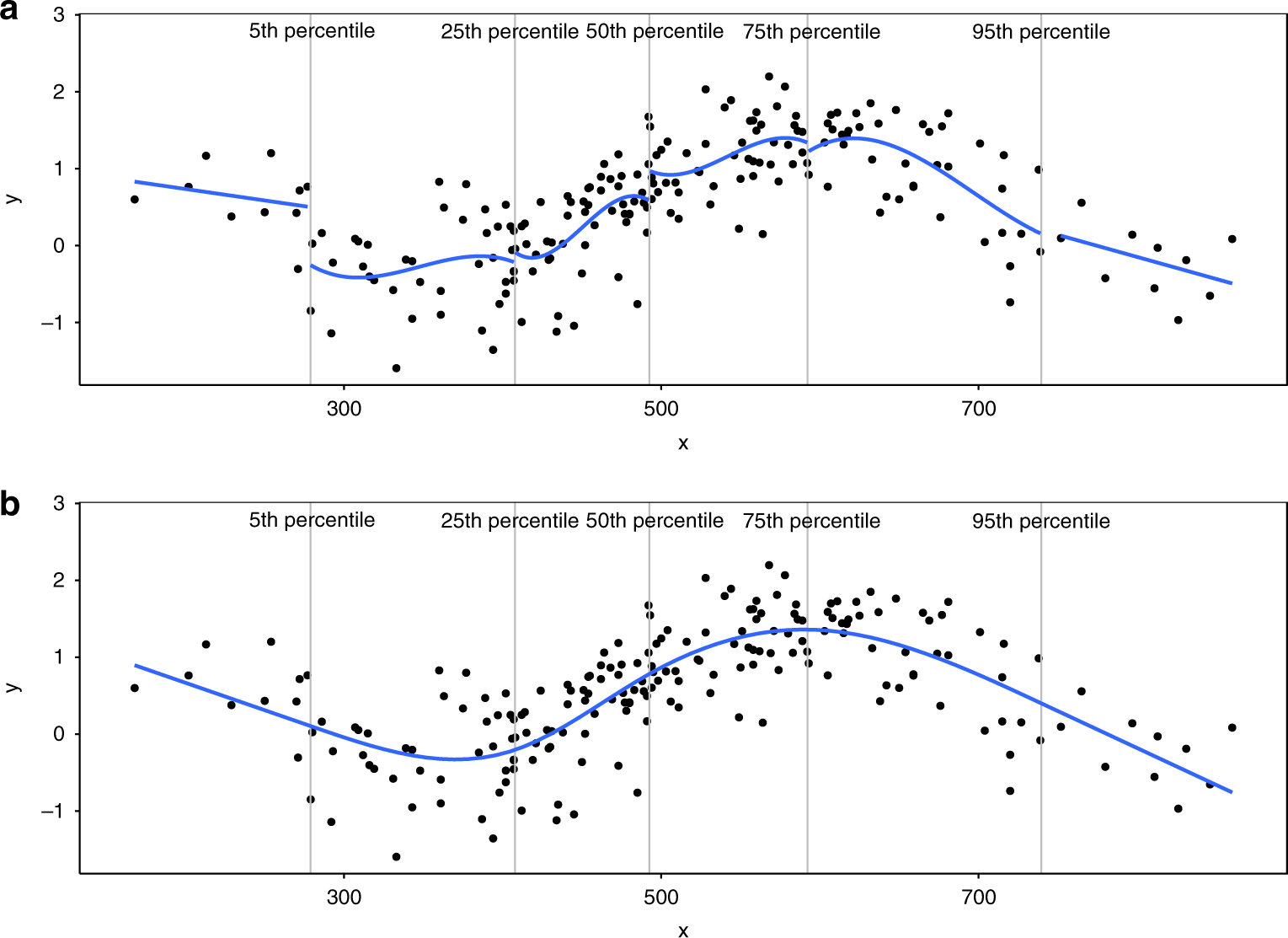
Natural cubic spline(ns)
Cubic + 처음과 끝은 Linear
- 처음보다 더 처음, 끝보다 더 끝(데이터에 없는 숫자)에 대한 보수적인 추정.
- 3차보다 1차가 변화량 적음.
Cubic spline in R
library(splines)
cs1 <- glm(time ~ bs(age, knots = c(40, 50, 60, 70)) + sex, data = colon)
cs2 <- glm(time ~ bs(age, df = 4) + sex, data = colon)
ns1 <- glm(time ~ ns(age, knots = c(40, 50, 60, 70)) + sex, data = colon)
ns2 <- glm(time ~ ns(age, df = 4) + sex, data = colon)
age.grid <- seq(min(colon$age), max(colon$age), by = 1)
with(colon, plot(age,time,col="grey",xlab="Age",ylab="Time"))
points(age.grid, predict(cs1, newdata = data.frame(age=age.grid, sex = 1)), col=1, lwd=1, type="l")
points(age.grid, predict(cs2, newdata = data.frame(age=age.grid, sex = 1)), col=2, lwd=2, type="l")
points(age.grid, predict(ns1, newdata = data.frame(age=age.grid, sex = 1)), col=3, lwd=3, type="l")
points(age.grid, predict(ns2, newdata = data.frame(age=age.grid, sex = 1)), col=4, lwd=4, type="l")
#adding cutpoints
abline(v = c(40, 50, 60, 70), lty=2, col="black")
legend("topleft", c("cs:knots" ,"cs:df", "ns:knots", "ns:df"), col = 1:4, lwd = 1:4)
Smoothing spline
mgcv R 패키지의 기본옵션.
Loess, Cubic spline
- Span, knot를 미리 지정: local 구간을 미리 지정.
Smoothing(penalized) spline
- 알아서.. 데이터가 말해주는 대로..
Penalized regression: Smoothing
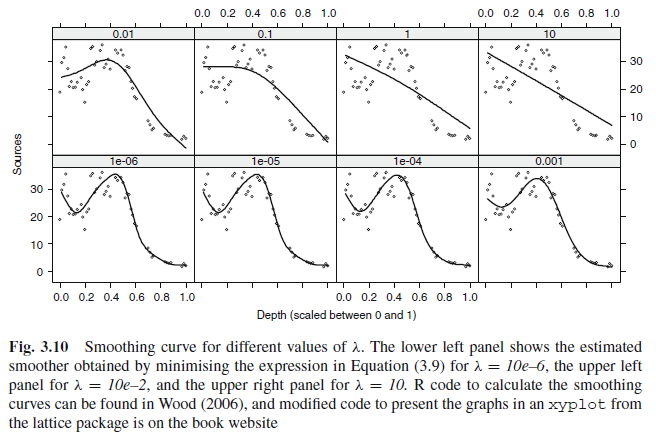
\[\begin{align*} \text{Minimize} ||Y - X\beta||^2 + \lambda \int f''(x)^2dx \end{align*}\]- \(\lambda\rightarrow 0\): 울퉁불퉁.
- \(\lambda\)가 커질수록 smoothing
- \(f''\) 는 기울기의 변화율, 제곱하면 절대값 의미
\(\lambda\)
Show gam result
- edf(Effective df): 자유도 - shrinkage penalty
Family: gaussian
Link function: identity
Formula:
time ~ s(age) + sex
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1533.194 31.619 48.489 <2e-16 ***
sex 8.354 43.851 0.191 0.849
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(age) 7.584 8.447 2.725 0.00437 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = 0.00938 Deviance explained = 1.4%
GCV = 8.9245e+05 Scale est. = 8.8784e+05 n = 1858 s(age)
0.04120719 Plot

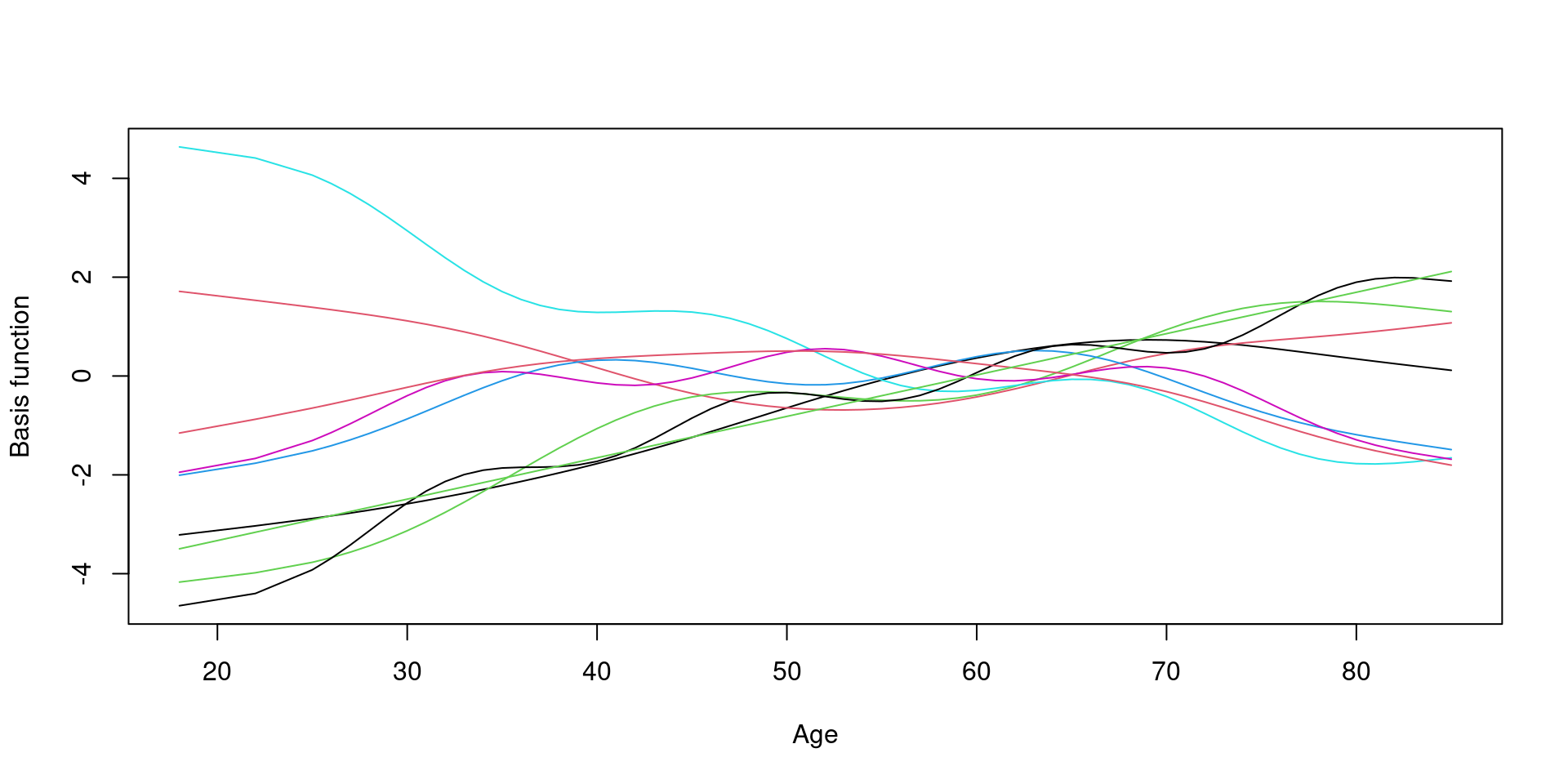
Basis function
Smoothing spline: basis function 들의 합
\[s(x) = \sum_{k = 1}^K \beta_k b_k(x)\]
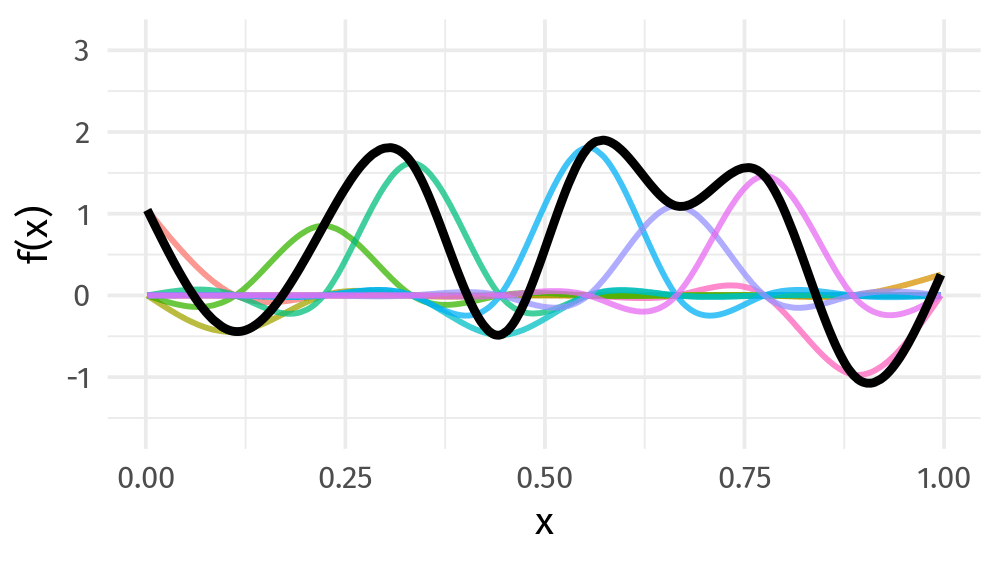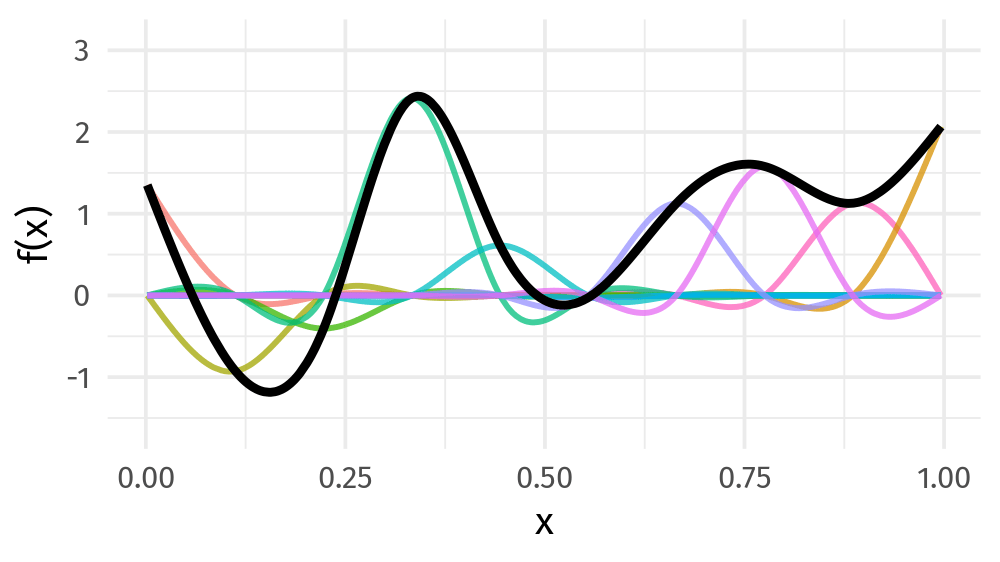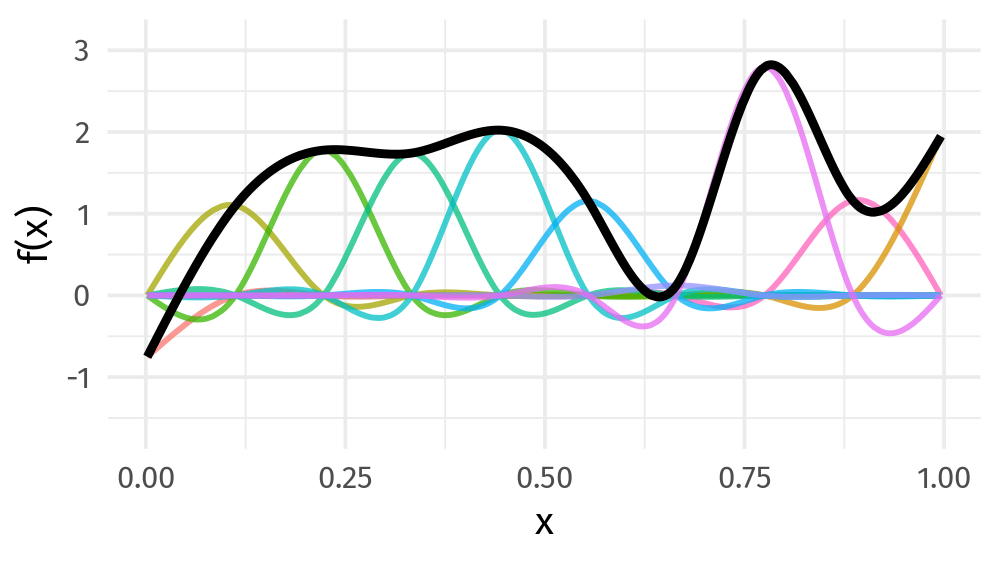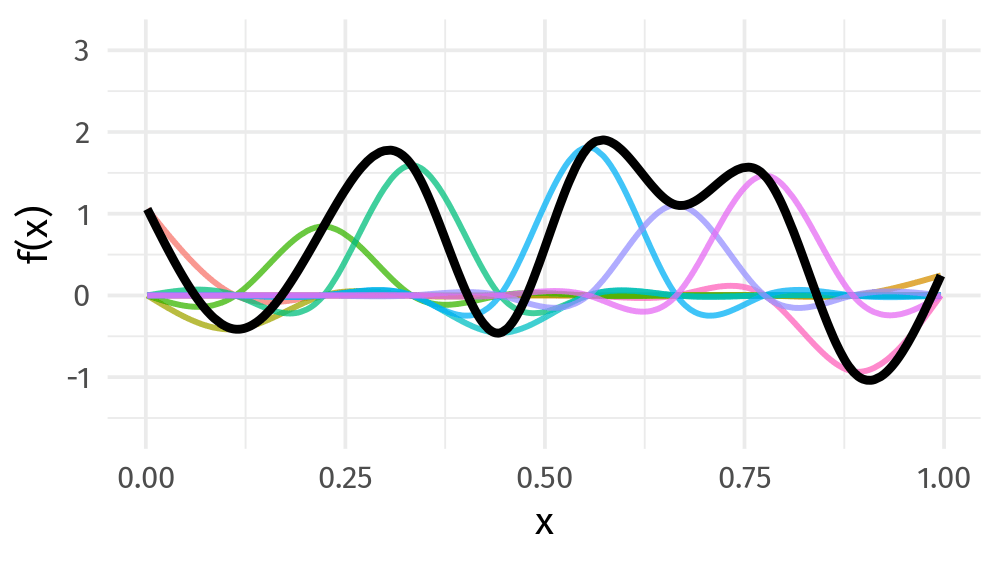
Basis function: value
Basis function: plot
9개의 basis function
GAM result
model_matrix 에 계수를 곱하면 곡선의 y값
(Intercept) sex s(age).1 s(age).2 s(age).3 s(age).4
1533.193607 8.353593 776.245594 1393.793067 232.785846 -1154.982151
s(age).5 s(age).6 s(age).7 s(age).8 s(age).9
-324.817013 -1018.225598 274.926370 3106.242791 -783.531680 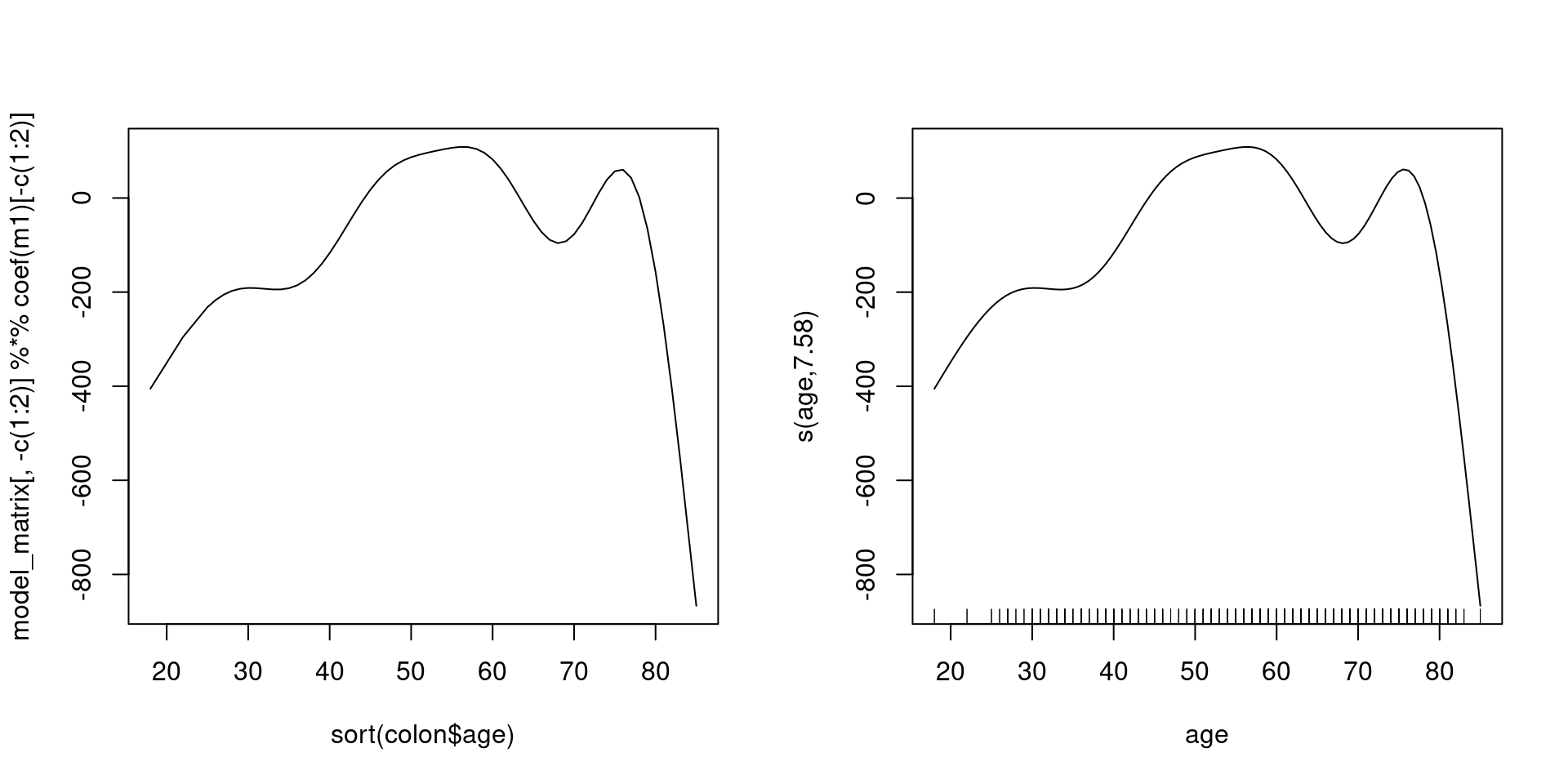
Change basis function
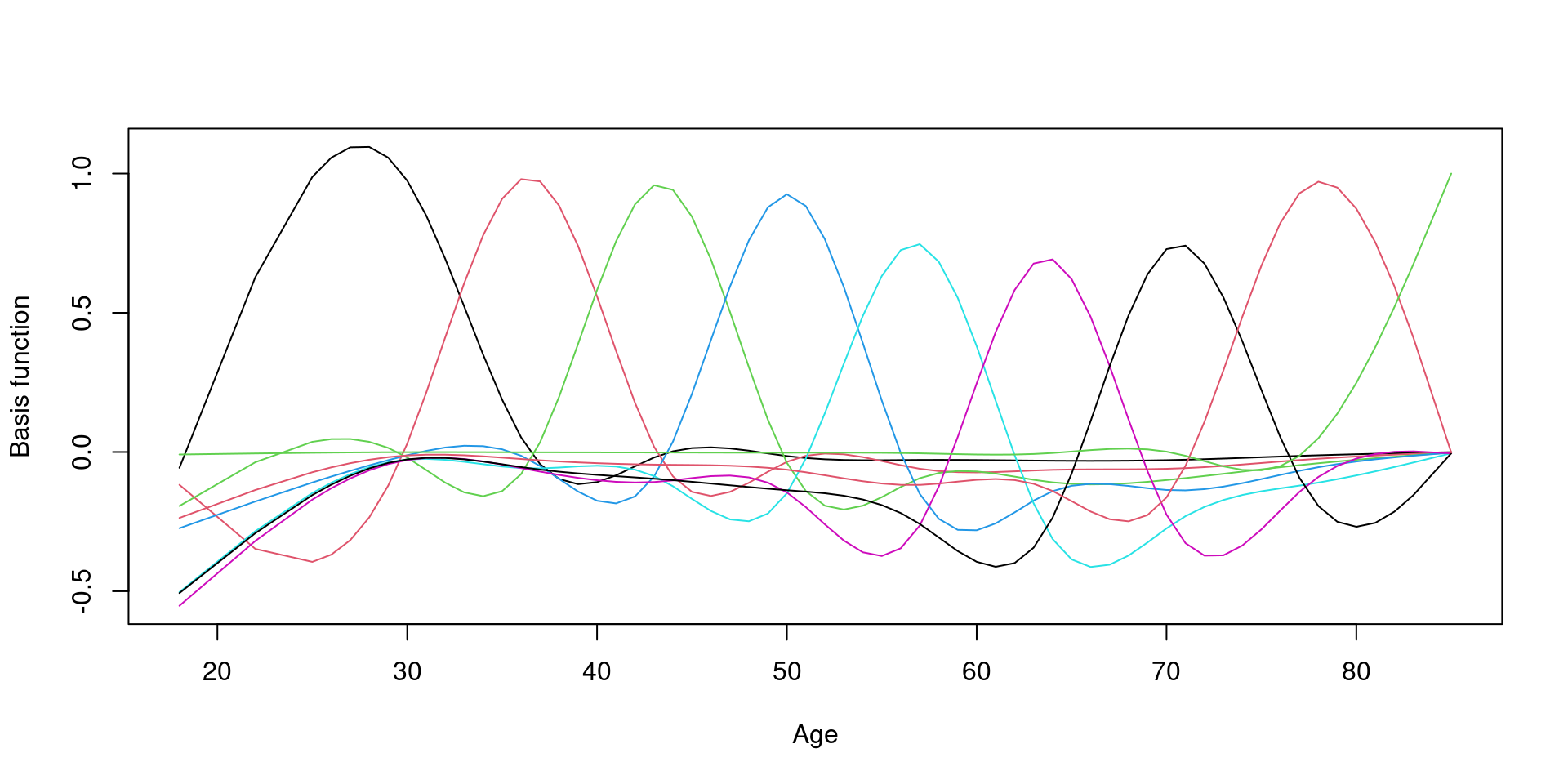
Restricted df
\(k = 6\): df의 최대값을 6으로 제한
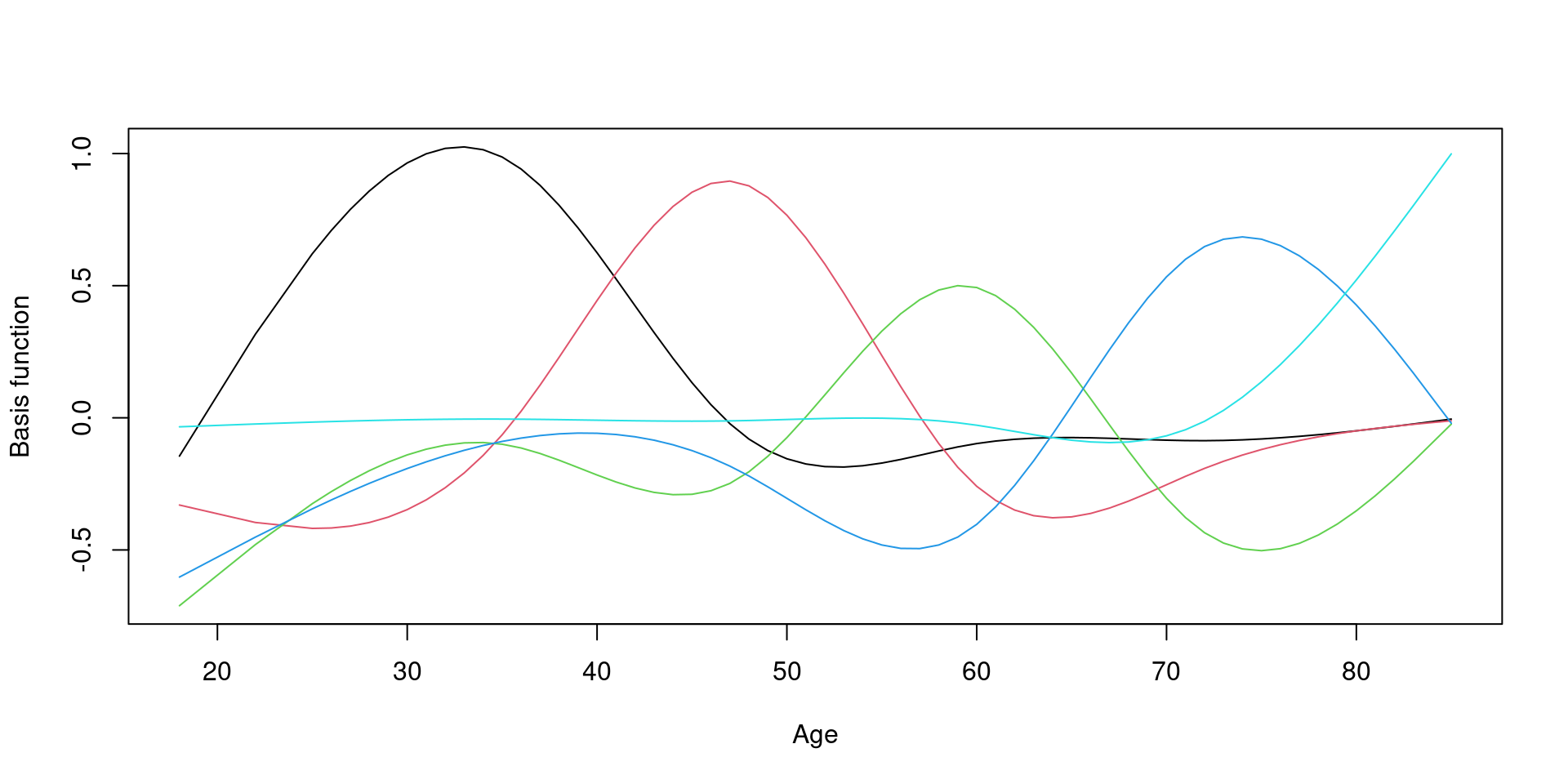
Fix \(\lambda\)
sp = 1000: \(\lambda\) 1000으로 고정, 거의 직선을 의미

Other distribution
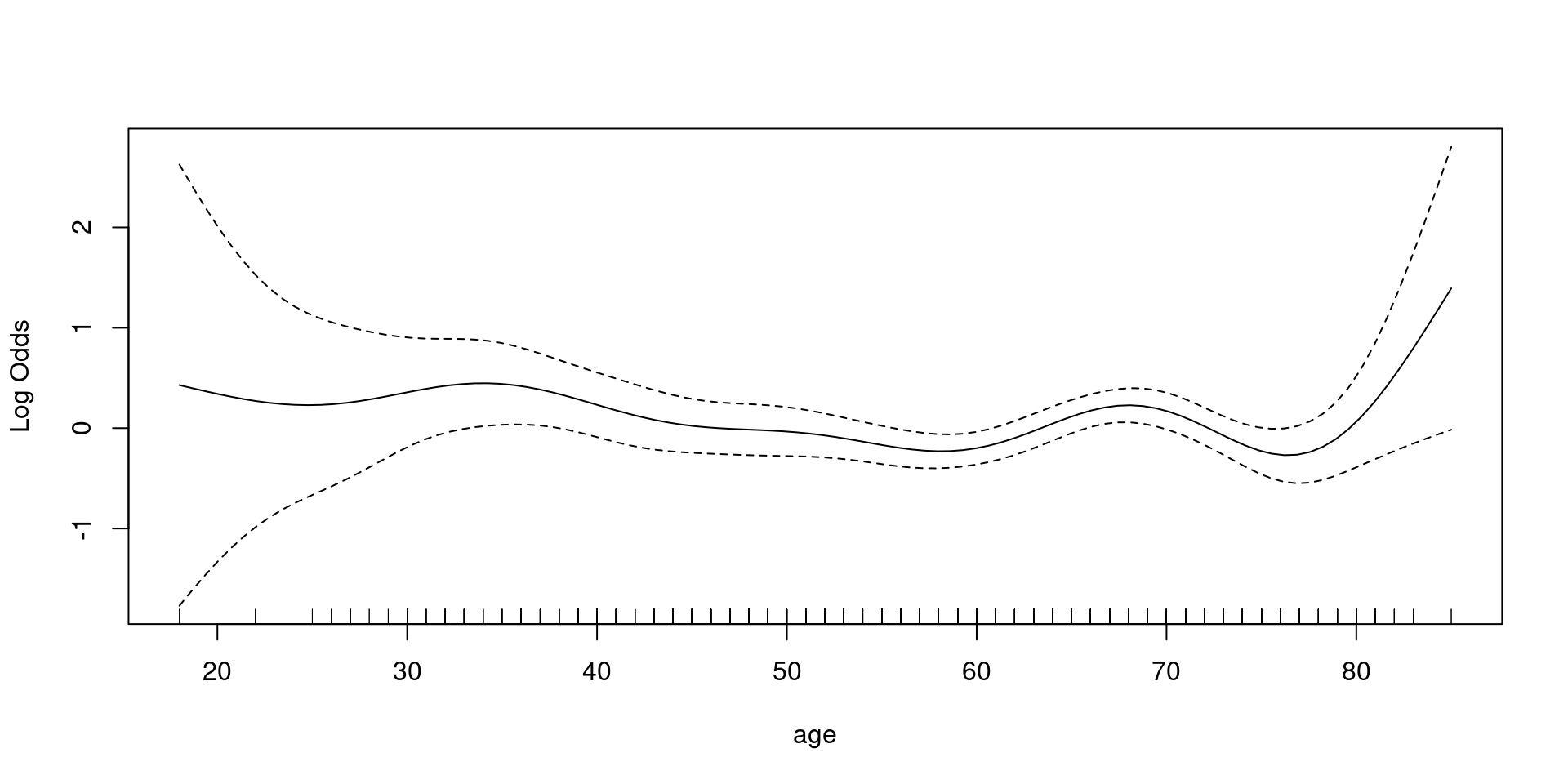
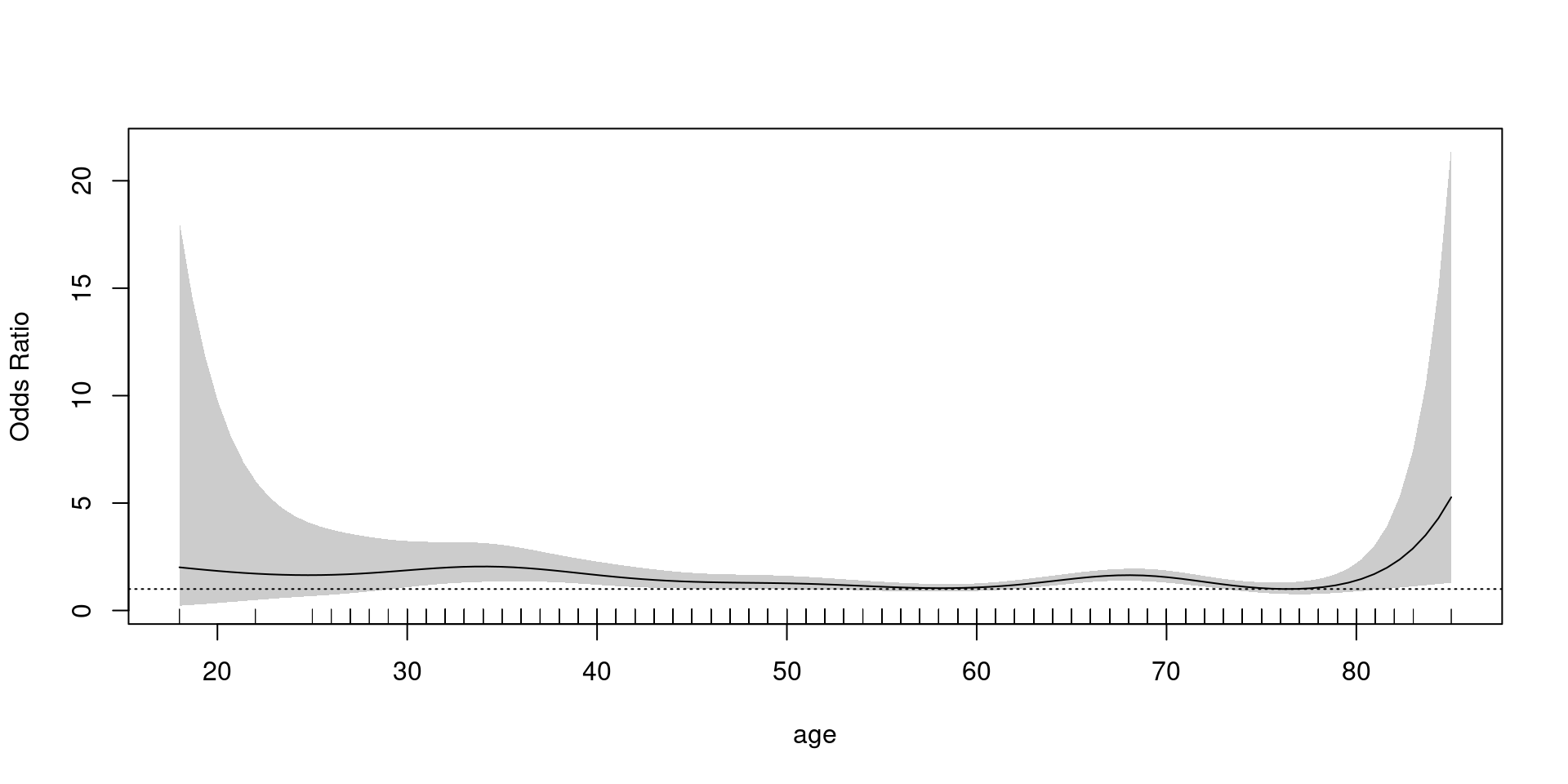
Logistic
family = binomial
- 해석에 주의: log Odds
- exponential transformation, reference(OR =1) 설정 필요
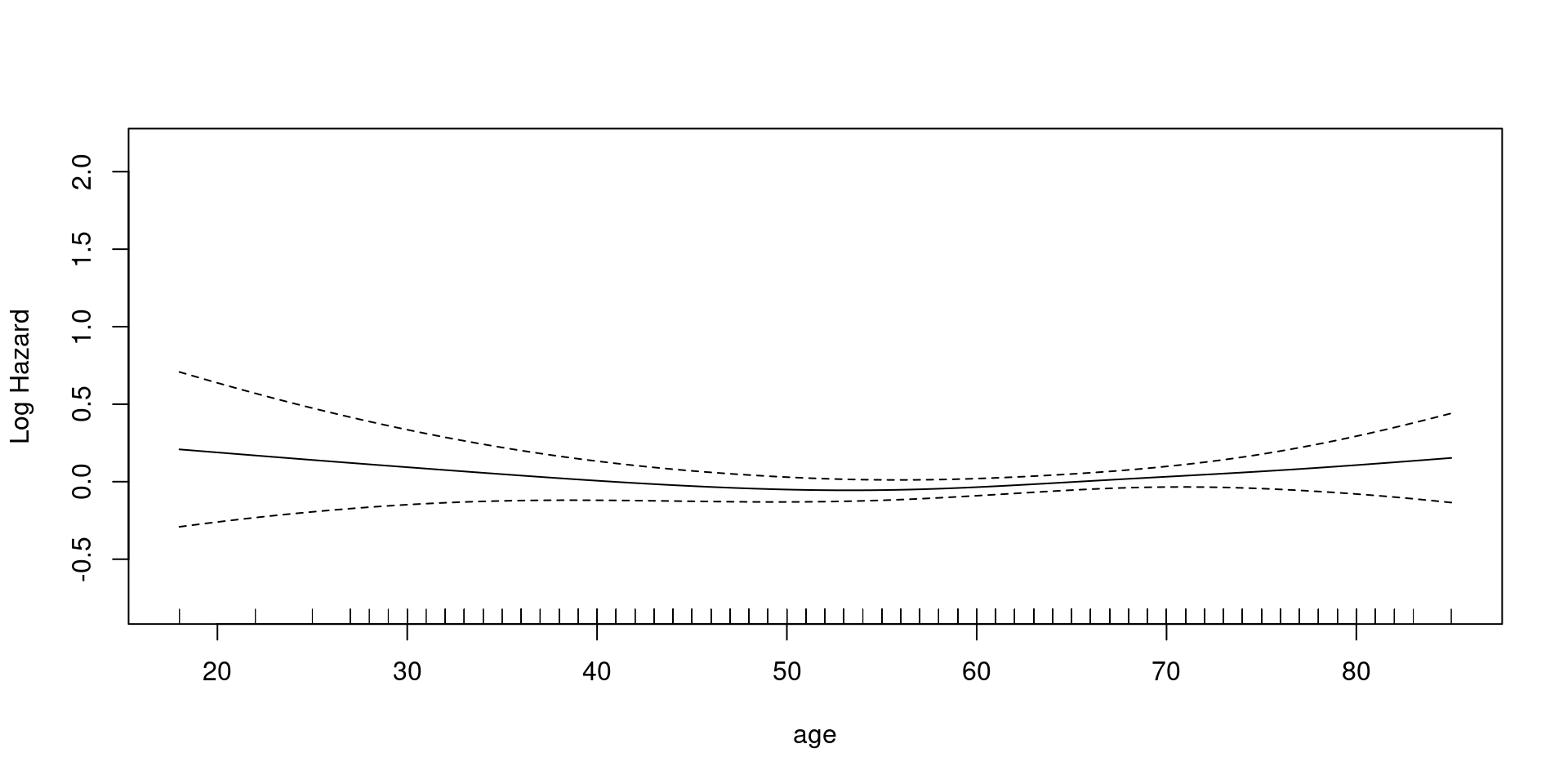
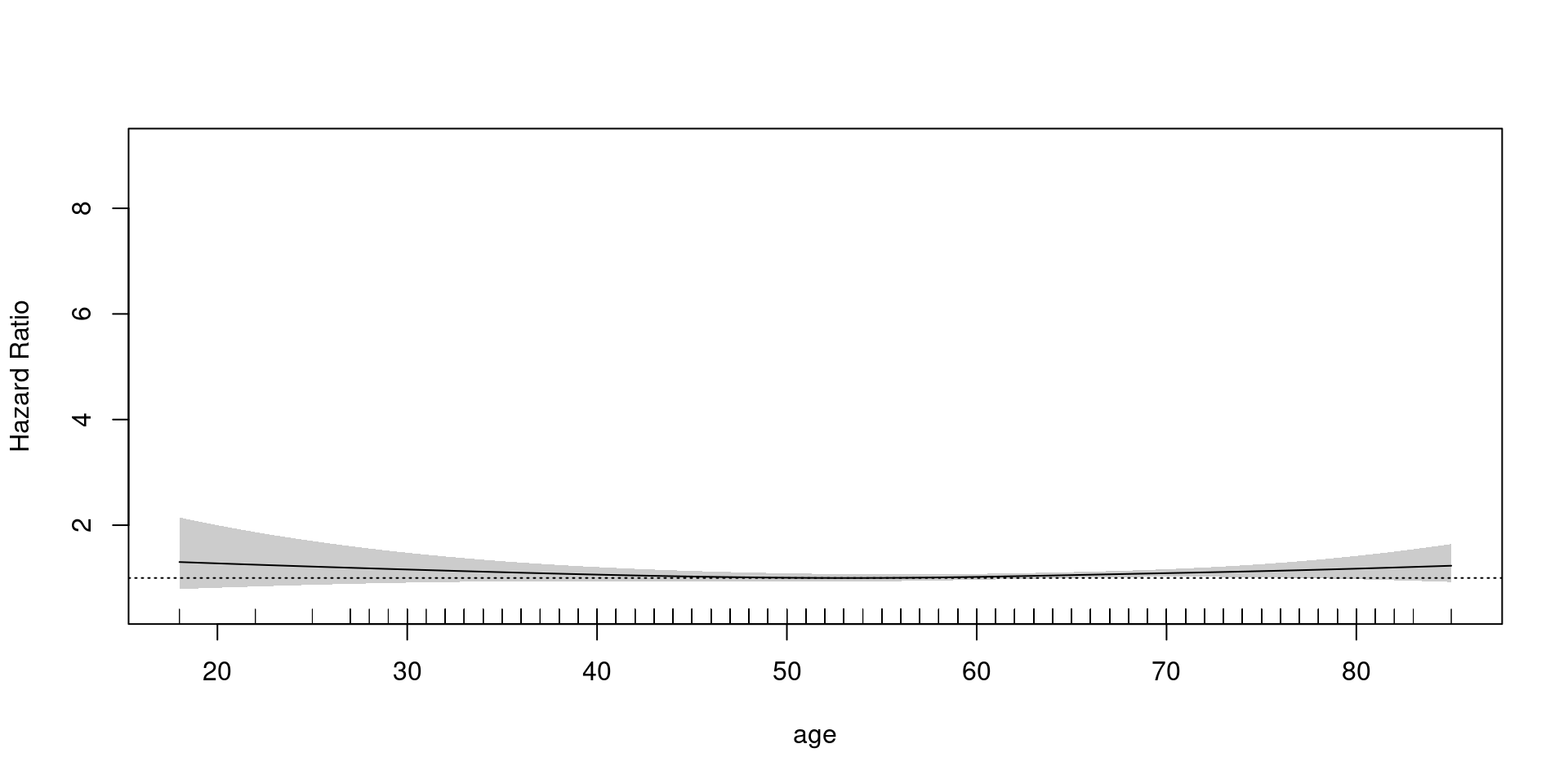
Coxph
family = cox.ph - weights = status
Poisson
family = poisson - exp trans
Quasi-poisson
Poisson 분포의 가정 평균=분산 이 만족하지 않을 때.
family = quasipoisson- \(평균 = \phi * 분산\)
- 곡선 자체는 그대로, 신뢰구간만 넓어짐(보수적 추정)
Thank you for your time

www.zarathu.com